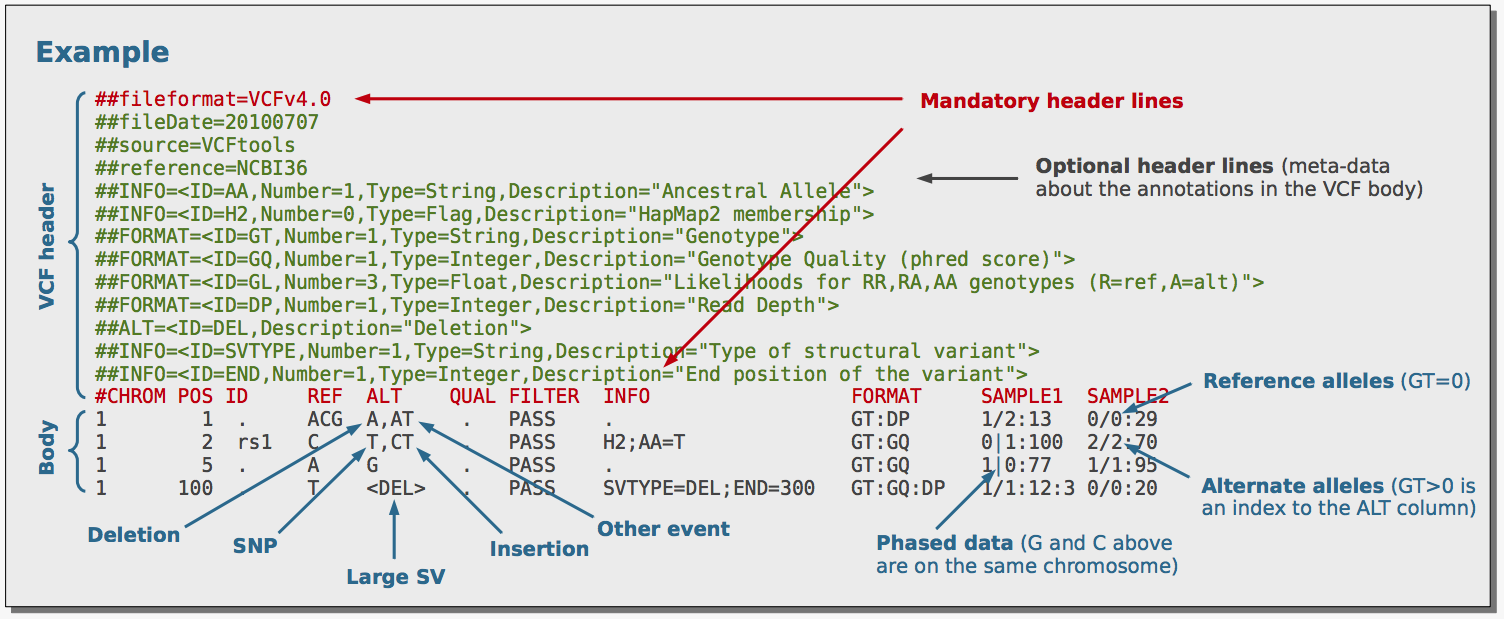
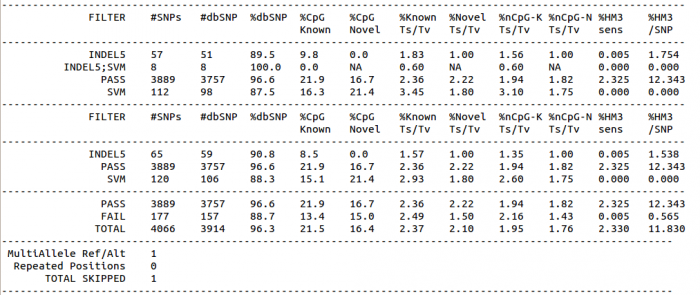
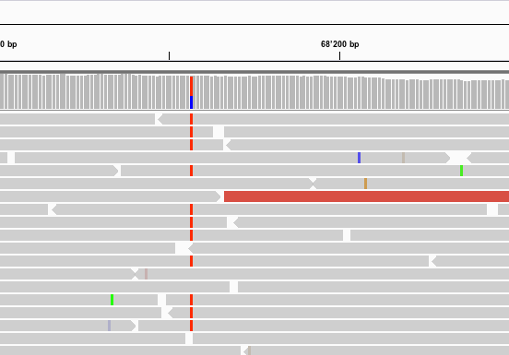
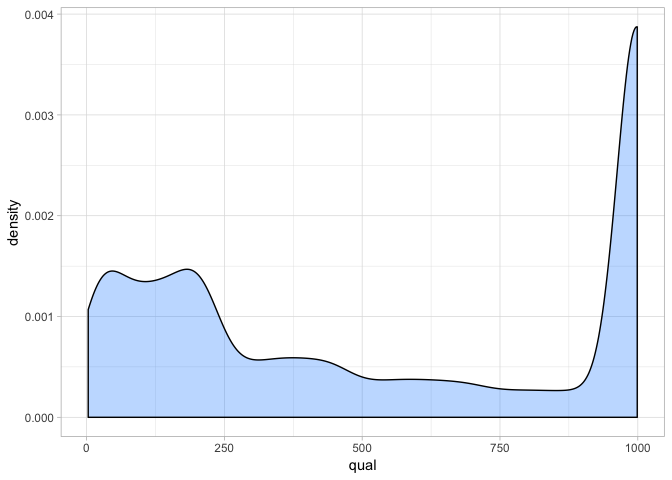
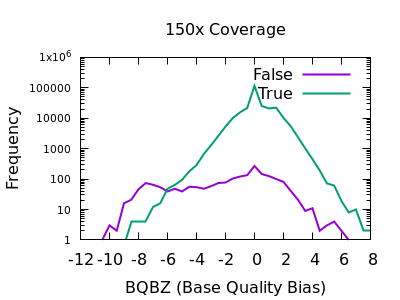
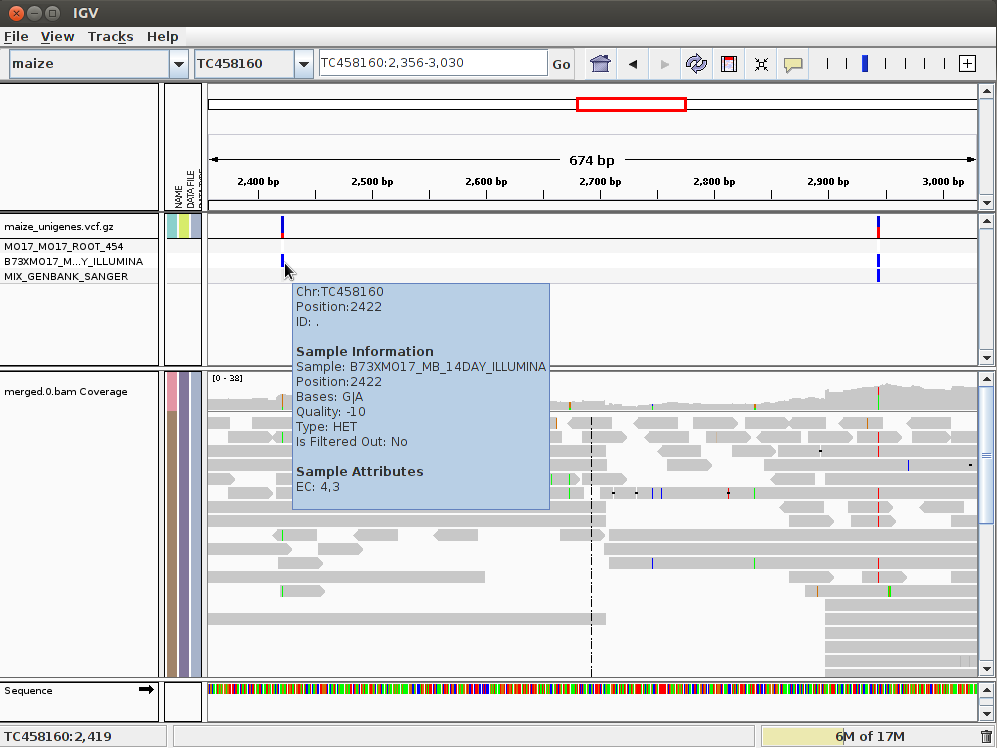
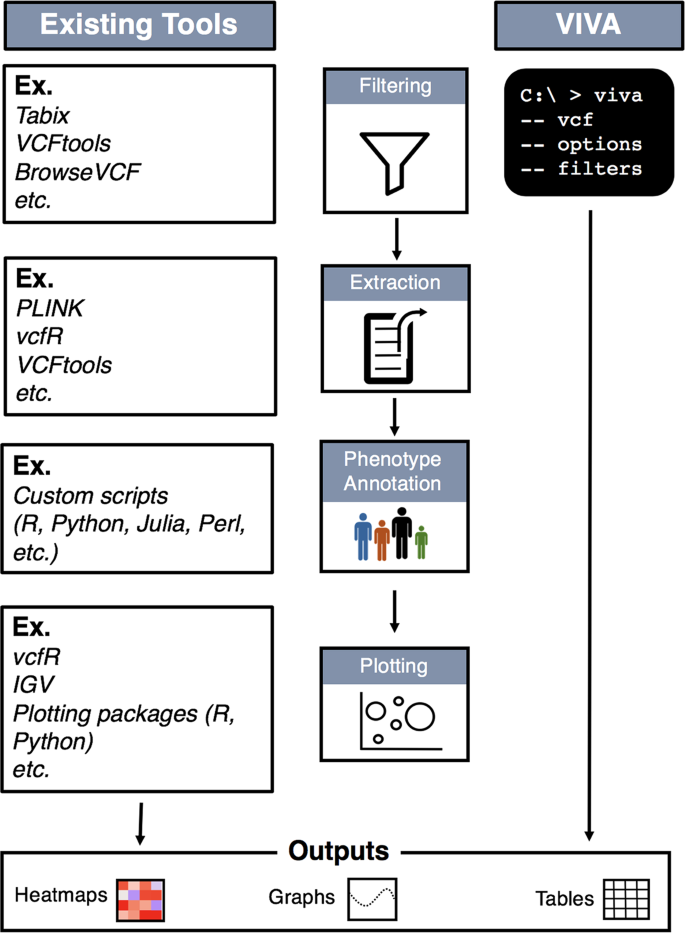
SNP markers tightly linked to root knot nematode resistance in grapevine (Vitis cinerea) identified by a genotyping-by-sequencing approach followed by Sequenom MassARRAY validation | PLOS ONE

snpfiltr: An R package for interactive and reproducible SNP filtering - DeRaad - 2022 - Molecular Ecology Resources - Wiley Online Library

snpfiltr: An R package for interactive and reproducible SNP filtering - DeRaad - 2022 - Molecular Ecology Resources - Wiley Online Library